|
| Rhinolophus malayanus, photo by Pipat Soisook |
by: Alexis M. Brown
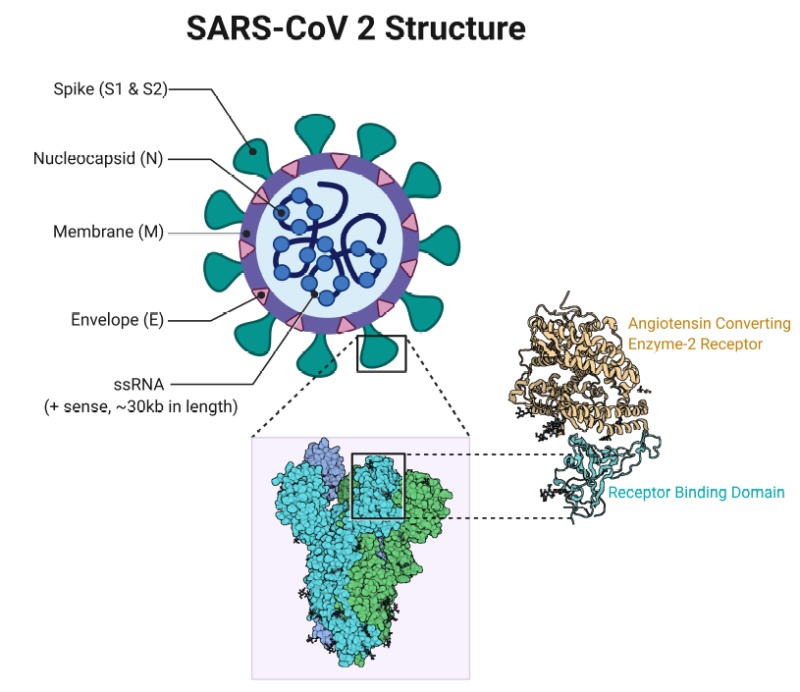
SARS-CoV-2, the coronavirus that causes COVID-19, shares a high degree of nucleotide similarity with a coronavirus of unknown human spillover potential isolated from bats. But does “high nucleotide similarity” between two viral strains mean that they are functionally equivalent, or that a virus found in wildlife has high spillover potential to humans? In order to answer these questions, we need to dive deeper into the interwoven stories of genome evolution, functional genes, and protein structure of both human SARS-CoV-2 and coronaviruses isolated from bats.
For SARS-CoV-2 to infect human cells, the virus uses the receptor-binding domain (RBD) of its spike (S) protein to match up with the angiotensin-converting enzyme 2 (ACE2) receptor on the surface of human cells. Once the virus uses its RBD “key” to gain access to the human cell, it can enter, utilize cellular machinery to replicate its RNA genetic material, and then proliferate to other human cells, eventually inducing an inflammatory immune response that causes acute disease symptoms known as COVID-19 (Figure 1). To understand where the human pathogen SARS-CoV-2 came from, we need to find a virus in wildlife populations that has both genetic and functional similarity to SARS-CoV-2, as well as the potential to be pathogenic to humans. As we find signatures of these features, we may be getting closer to figuring out what the “parent” strain of the SARS-CoV-2 was and the animal reservoir from which it emerged.
|
|
||||
|
Figure 1. Relationship between SARS-CoV-2 coronavirus, RBD of viral Spike protein, and human ACE2 receptors. ACE2 receptors block entry into human cells, unless the agent seeking entry has the appropriate protein “key”. The RBD (right) of the SARS-CoV-2 Spike protein is this key, which matches perfectly to ACE2, binds the virus to the receptor, and allows successful entry of the virus into the human cell. The virus will then use cellular machinery to replicate and infect other healthy cells. If any mutation or genetic recombination occurring within the SARS-CoV-2 virus results in structural changes to the RBD, then the virus may not infect human cells. Sources: Cascella et al. (2020) (left), Sriram et al. (2020) (right).
|
|||||
Previously, the closest we’ve come to identifying a potential source of SARS-CoV-2 has been the isolation of coronavirus RaTG13 from Rhinolophus affinis in China. The RaTG13 virus shows 96% total nucleotide identity to SARS-CoV-2, and 93% nucleotide identity in the S-protein functional gene. However, when we look more closely at the key aminoacids involved interacting with the human ACE2 protein in the RBD region of the S-protein, RaTG13 only shares identity with 1 out of the 5 amino acid residues present in the RBD region of SARS-CoV-2. This means the protein sequence in the bat virus RBD required for gaining entry to human cells via ACE2 receptors is actually not very similar to that of the human virus. If the protein sequence results in structural changes in the actual shape of the protein, this would mean that the virus cannot bind to human ACE2 receptors and thus cannot infect human cells. If the bat virus can’t infect human cells, it is unlikely to be pathogenic to humans and unlikely to be the direct parent of SARS-CoV-2.
Based on previous outbreaks caused by coronaviruses, SARS and MERS, one of the current hypotheses posits the need for an intermediate host in which viruses from a bat reservoir could have mutated and/or recombined to become functionally similar to SARS-CoV-2, jumping into human hosts from the intermediate instead of directly from the bat hosts. For SARS-CoV-2, Zhang et al. (2020) found that the RBD of a coronavirus isolated from the Malayan pangolin (Manis javanica) shows higher amino acid sequence similarity to SARS-CoV-2 than RaTG13 does, and implied that pangolins could be the intermediate host in which the “parent” bat virus mutated to form SARS-CoV-2. We have already established that the bat coronavirus RaTG13, hypothesized as being a potential “parent virus” to SARS-CoV-2, is unlikely to be capable of infecting human cells due to the dissimilarity in the two virus’ RBD functional regions. In order for SARS-CoV-2 to have gained the functionality to infect human cells, it would have to have shuffled, or recombined, its genome. It could gain the ability to infect humans either by a) recombining its own genome in situ within the reservoir host, or b) hybridizing and recombining with a new viral strain that has the appropriate functional domains in an intermediate host, like the pangolin.
A new study, however, has described a new bat coronavirus that shows evidence that the parent virus of SARS-CoV-2 may not have required an intermediate host to incorporate humans as a new host. The novel virus, RmYN02, was isolated from Rhinolophus malayanus. It shows lower sequence identity to SARS-CoV-2 than RaTG13, both in its whole genome (93.3%) and the S-protein gene (71%). But Zhou and colleagues found something very interesting when they looked for signatures of natural in situ recombination of bat coronaviruses. SARS-CoV-2 is characterized by a unique four amino-acid insertion at a specific site of the S-protein. In RmYN02, there is a three amino-acid insertion at this exact site in its own S-protein (Figure 2). Although the insertion in RmYN02 contains three different amino acids than those found in the SARS-CoV-2 insertion, its presence in a sample from a wild bat is an important finding: SARS-CoV-2 shows patterns of natural recombination events occurring in the virus without requiring an intermediate host. If this inference about recombination is correct, then bat viruses may not require intermediate hosts between bats and humans to undergo recombination in their functional genes, leading to potentially adaptive structural changes and allowing SARS-CoV-2 to infect human cells.
|
Figure 2. Amino acid residue similarity in S-protein RBDs between SARS-CoV-2 and 8 wildlife coronaviruses. Note that only RmYN02 (bat coronavirus isolated from R. malayanus) and SARS-CoV-2 show amino acid insertions at the polybasic cleavage site. The authors suggested this is evidence for in-situ recombination events frequently occurring in bat coronaviruses, which may imply that SARS-CoV-2’s parent virus did not need an intermediate host in order to gain the adaptive functional changes required to infect human hosts. Figure from Zhou et al. (2020).
|
We are learning more about the relationships between SARS-CoV-2, bat coronaviruses, and other wildlife coronaviruses every day, but much more research is needed. So much is still currently unknown, specifically regarding the wildlife reservoir of SARS-CoV-2, the pathogenic potential of bat coronaviruses to humans, and the adaptive recombination potential of wildlife viruses. We are still unsure of the details regarding the transmission pathway of SARS-CoV-2: Genetic similarity points to bats as being the reservoir source, but did the bat virus recombine in situ and directly spill over to humans? Was there an intermediate host? How do all these players come together in the picture of the reservoir/intermediate host/human transmission pathway?
The most important takeaway from this new knowledge? Viruses are naturally occurring components of ecosystems around the world, and have evolved in tandem with their wildlife reservoirs. Most wildlife viruses are unlikely to be harmful to humans. However, as humans continue to alter the natural world and destroy wildlife habitats, we shrink the physical barriers between wildlife and human populations. By shrinking these barriers, we risk exposing human populations to the small percentage of wildlife viruses that do have the potential to be harmful to humans by amplifying human-wildlife contact that would otherwise be minimal. Moving forward, our approach to preventing zoonotic outbreaks, must focus around minimizing our role in wildlife-harming activities that increase the likelihood of viral spillover. Without active efforts to change the ways in which we interact with wildlife and ecosystems, future pandemics may be inevitable.
References and Further Readings
- Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T.S., Herrler, G., Wu, N.-H., Nitsche, A., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181, 271-280.e8.
- Stevens, C. (2020). The Origins of SARS-CoV-2: Part 2. Accessed 21 June 2020.
- Zhang, T., Wu, Q., and Zhang, Z. (2020). Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol. 30, 1346-1351.e2.
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273.
- Zhou, H., Chen, X., Hu, T., Li, J., Song, H., Liu, Y., Wang, P., Liu, D., Yang, J., Holmes, E.C., et al. (2020). A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol. 30, 2196-2203.e3.